Abstract
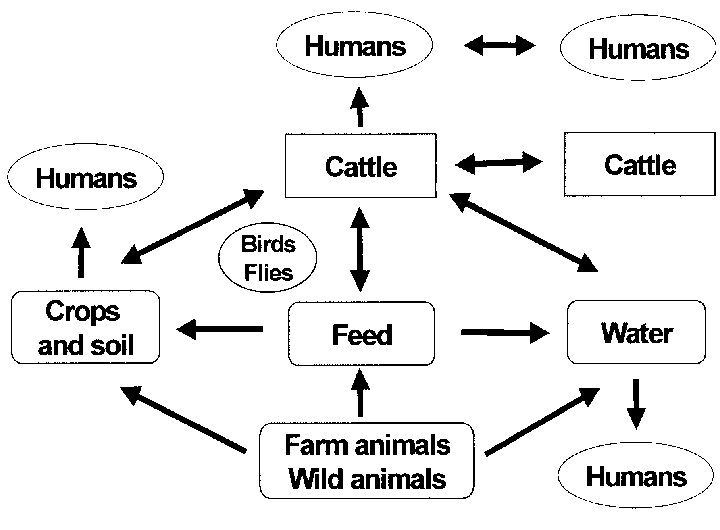
There are 29 E. coli genomic sequences available, mostly related to studies of species diversity or mode of pathogenicity, including two genomes of the known clone O157:H7. However, genome studies of closely related clones intended to expose the details of evolutionary change have not been performed. Here we sequence the genome of a strain O55:H7, closely related to the major pathogenic clone O157:H7, with published genomic sequences, and perform comparative genomic and proteomic analysis. We were able to assign most of the differences between the genomes to single mutations, recombination events, or lateral gene transfer events, in specific lineages.
Major differences include a type II secretion system present only on chromosome O55:H7, fewer type III secretion system effectors on O55:H7, and 19 phage genomes or phage-like elements on O55:H7 compared to 23 in O157:H7, with only three common to both. Many other changes were found in the O55:H7 and O157:H7 lineages, but overall there have been more changes in the Escherichia coli O157:H7 Recombinant lineages. For example, we found 50% more synonymous mutational substitutions in O157:H7 compared to O55:H7. The two strains also diverged at the proteomic level.
Synonymous mutational SNPs were used to estimate a divergence time of 400 years using a new clock rate, in contrast to 14,000 to 70,000 years using traditional clock rates. The same approaches were applied to three closely related extraintestinal pathogenic E. coli genomes, and similar levels of mutation and recombination were found. This study revealed for the first time the full range of events involved in the evolution of clone O157:H7 from its ancestor O55:H7 and suggested that O157:H7 arose recently. Our findings also suggest that E. coli has a much lower frequency of recombination relative to mutation than was observed in a comparative study of a Vibrio cholerae lineage.
Purity: greater than 90% as determined by SDS-PAGE.
Destination names: ompC
Uniprot No.: Q8XE41
Research Area: Microbiology
Alternative Names
ompC; Z3473; ECs3104; outer membrane porin C; outer membrane protein 1B; outer membrane protein C; Porin OmpC
Species: Escherichia coli O157:H7
Source: E.coli
Expression Region: 22-367aa
Mole Weight: 58.4 kDa
Protein length: Total length of the mature protein
Tag information: N-terminal 10xHis-SUMO-tagged and C-terminal Myc-tagged
Form: Liquid or Lyophilized Powder
Note: We will preferably ship the format we have in stock, however, if you have any special requirements for the format, please remark your requirement when placing the order, we will prepare according to your demand.
Buffer
If the dosage form is liquid, the default storage buffer is Tris/PBS based buffer, 5%-50% glycerol.
Note: If you have any special requirements for glycerol content, please remark it when you order. If the administration form is a lyophilized powder, the buffer before lyophilization is Tris/PBS-based buffer, 6% trehalose, pH 8.0.
Reconstitution
We recommend that this vial be briefly centrifuged before opening to bring the contents to the bottom. Reconstitute protein in sterile deionized water at a concentration of 0.1-1.0 mg/mL. We recommend adding 5-50% glycerol (final concentration) and an aliquot for long-term storage at -20°C/-80°C. Our final default glycerol concentration is 50%. Customers could use it for reference.
Storage Conditions
Store at -20°C/-80°C upon receipt, need to be aliquoted for multiple uses. Avoid repeated cycles of freezing and thawing.
Shelf life
Shelf life is related to many factors, storage condition, buffer ingredients, storage temperature and the stability of the protein itself. Generally, the shelf life of the liquid form is 6 months at -20°C/-80°C. The shelf life of the lyophilized form is 12 months at -20°C/-80°C.
Delivery time: 3-7 business days
Notes: Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
Growth conditions for proteomic analysis
E. coli O55:H7 strain CB9615 and O157:H7 strain EDL933 were grown to early exponential phase and early stationary phase in 100 ml of Dulbecco’s modified Eagle’s medium (DMEM, Gibco™, Invitrogen corporation). Bacterial growth was monitored by measuring the absorbance of light at 600nm using a 752 UV visible spectrophotometer at 1h intervals for 8h. Cells were collected by centrifugation (8000 xg for 10 min at 4°C). For each condition, proteins prepared from three independent cultures were pooled for further analysis.
Protein extraction for 2-DE
To extract cytosolic proteins, harvested cells were washed twice in 50 mL wash buffer (0.01 M Tris, 0.25 M sucrose, pH 7.2) and then suspended in lysis buffer (8 M urea). , 2M thiourea, 4% CHAPS, 1% DTT, 0.8% ampholine pH 3-10) followed by sonication for 5 min using an Ultraschallprozessor UP200S (Hielscher). Insoluble material was removed by centrifugation at 25,000 xg for 1 hour at 4°C. The supernatant was collected and stored at -80°C until use. Protein concentration was determined by the Bradford method with bovine serum albumin as standard.
Gel digestion
Protein spots were manually excised from Coomassie Brilliant Blue G-250 (BBI) stained gels. The excised gel spots were washed several times with destaining solutions (deionized water for 15 min three times and then with 50% (v/v) acetonitrile containing 25 mM NH4HCO3 for 15 min three times), then with pure water for 15 min. min three times. Gel spots were dehydrated in 50ul 100% ACN for 20 min at room temperature and rehydrated in 25mM NH4HCO3 containing 0.005ug/ul modified trypsin (Promega). Proteins were digested with trypsin for 16 to 20 hours at 37°C to generate peptides.
MALDI-TOF analysis
Peptides were analyzed by MALDI-TOF using a 4700 series proteomics analyzer (Applied Biosystems). Internal calibration was obtained with the peptides derived from 700–4000 Da. CHCA (sigma) was used as a matrix. The spectra peak list was created by the Peak-to-MASCOT script of the 4700 Explorer software. The samples were analyzed by PMF and compared with a local database using the pet algorithm of the GPS software.
Data analysis
To identify proteins with significant differences in protein synthesis between the early stationary phase and the exponential phase, we determined the ratios of the total normalized amounts of the corresponding spots in the images of the two samples. Spots with synthesis ratios of >2.0 (top) and <0.5 (bottom) in all 3 replicates were considered to contain proteins with significant changes in synthesis rate.